Automated clean-up of pesticides in oats using ISOLUTE® cSPE for QuEChERS
By Biotage
QuEChERS is a sample preparation technique commonly used in food safety testing for pesticide residue analysis. This two-stage extraction and clean-up technique was developed to streamline sample processing; making it Quick, Easy, Cheap, Effective, Rugged and Safe. ISOLUTE® cSPE for QuEChERS are pre-packed cartridges designed to further improve the efficiency of the QuEChERS clean-up procedure.
Traditionally, clean-up of QuEChERS extracts for pesticide residue analysis relies on dispersive solid phase extraction (dSPE). The dSPE workflow requires that the sample is shaken with the dSPE media and then centrifuged to separate the clean sample extract. Alternatively, ISOLUTE®cSPE for QuEChERS cartridges are packed with QuEChERS ‘dispersive SPE’ sorbent blends allowing convenient flow-through sample processing using column solid phase extraction (cSPE). In addition, the cSPE workflow can be automated on a Biotage® Extrahera™ sample preparation workstation leading to increased throughput, reduced errors, and improved data quality.
 Figure 1. Example of pesticide structures.
Figure 1. Example of pesticide structures.
Introduction
This application note describes the extraction of a panel of 77 commonly analyzed pesticides from oats using a two-step QuEChERS workflow. Samples are first homogenized and extracted with ISOLUTE® EN QuEChERS extraction salts using the Biotage® Lysera™ bead mill homogenizer. Following centrifu- gation, an aliquot of extract is cleaned-up using ISOLUTE® cSPE for QuEChERS cartridges and processed using a Biotage® Extrahera™ automated sample preparation workstation. Extracts are evaporated using TurboVap® Dual 96 and analysis is performed using GC-MS.
This application note includes optimised conditions for homog- enization and extraction, automated clean-up using ISOLUTE® cSPE for QuEChERS cartridges, evaporation and analysis.
Analytes
To demonstrate the versatility of the sample preparation approach, a panel of 77 analytes were selected to reflect a broad range of pesticide classes and chemical characteristics. The pesticides analyzed in this application note are listed below.
Carbofuran 1, Propamocarb, Mevinphos, Acephate, Captan, Carbaryl 1, Methiocarb, Propoxur, Ethoprophos, Dimethoate, Carbofuran 2, Atrazine, Quintozene, Diazinon, Pyrimethanil, Parathion-methyl, Metalaxyl, Carbaryl 2, Pirimiphos-methyl, Malathion, Chlorpyrifos, Oxamyl, Cyprodinil, Thiabendazole, Fluopyram, Fipronil, Chlordane, Prallethrin 1, Prallethrin 2, Paclobutrazol, Imazalil, Isoprothiolane, Tricyclazole, Fludioxonil, Buprofezin, Myclobutanil, Trifloxystrobin, Cyflufenamid, Chlorfenapyr, Penthiopyrad, Quinoxyfen, Propiconazole, Fenhexamid, DDT, Kresoxim-methyl, Fluopicolide, Tebuconazole, Propargite, Piperonyl butoxide, Iprodione, Fenoxycarb, Acetamiprid, Bifenthrin, Fluxapyroxad, Bifenazate, Fenpropathrin, Fenamidone, Pyriproxyfen, Lambda- Cyhalothrin, Ametoctradin, Permethrin 1, Pyridaben, Permethrin 2, Coumaphos, Fenbuconazole, Cyfluthrin 1, Cyfluthrin 2, Boscalid, Cypermethrin 1, Cypermethrin 2, Cypermethrin 3, Etofenprox, Difenoconazole, Indoxacarb, Azoxystrobin, Famoxadone, Dimethomorph.
Sample preparation procedure
Format
Step 1: QuEChERS Extraction: ISOLUTE® EN QuEChERS extrac- tion salts (4 g MgSO4, 1 g sodium citrate, 0.5 g sodium citrate sesquihydrate + 1 g sodium chloride) p/n Q0020-15V
Step 2: cSPE Clean-up: ISOLUTE® EN General 200 mg/3 mL (Tabless), p/n Q0035-0020-BG
Homogenization
10 g of oats were weighed into a 50 mL centrifuge tube and 10 mL of water was added to the oats. 3.3 g of metal beads were added to the centrifuge tube and three samples were homog- enized simultaneously using the Biotage® Lysera and 50 mL tube carriage with the following settings:
- Speed: 5 m/s
- Processing Time: 15 seconds
- Number of Cycles: 2
- Dwell Time: 10 seconds
Step 1: QuEChERS extraction
10 mL of acetonitrile and the ISOLUTE® EN QuEChERS extraction salts (comprising of 4 g MgSO4, 1 g sodium citrate, 0.5 g sodium citrate sesquihydrate and 1 g sodium chloride) were added to the homogenized oats and shaken using the Lysera with the following settings:
- Speed: 2.4 m/s
- Processing Time: 15 seconds
- Number of Cycles: 1
- Dwell Time: N/A
The samples were then centrifuged for 5 minutes at 5000 RCF. 1 mL of the supernatant was utilized for subsequent cSPE clean-up.
In order to evaluate the cSPE for QuEChERS clean-up, the supernatant was spiked at a concentration of 96 ppb with a mixture of 77 pesticides (see Additional Information). No internal standards were used.
Step 2: Automated clean-up procedure
An excess of 1 mL of supernatant from step 1 was transferred into 12 x 75 mm test tubes and placed into the Biotage® Extrahera™ Classic, (position 4, sample rack 12 x 75 mm, 24 positions). ISOLUTE® EN General 200 mg/3 mL (Tabless) (p/n Q0035-0020-BG) were placed onto the Extrahera™ Classic (position 3, column rack 24 x 3 mL), and collection vials were placed in position B (collection rack 12 x 75 mm, equipped with rack inserts). Using the Extrahera™, 1 mL of QuEChERS extract was loaded onto each column. The columns were processed automatically by applying pressure (0.6 bar for 210 seconds, followed by 5 bar for 15 seconds and a 15 second plate dry). Extracts were collected into high recovery microsampling GC vials.
For options to increase batch size and throughput with minimal sample handling steps, see Appendix 2.
Post extraction
Samples were evaporated using the TurboVap® 96 Dual with the following parameters:
- 24 Configuration, Single Mode
- Gas Temp: 40 °C
- Plate Temp: 40 °C
- Gas Flow: 25 L/min
- Plate Height: 56 mm
- Time: 30 minutes
Extracts were reconstituted with 30 µL of toluene (added directly to the GC vial) and vortexed prior to analysis.
Analytical conditions
Table 1. GC conditions
|
Instrument |
Agilent 7890A GC |
|
Column |
Restek Rtx-5MS 30 m x 0.25 mm ID x 0.25 µm |
|
Carrier gas |
Helium 1.2 mL/min |
|
Inlet |
Splitless, 280°C, 50 mL/min at 1 min |
|
Injection volume |
1.3 µL |
|
Wash solvent |
Methanol |
|
Oven settings |
Available on request |
|
Transfer line |
300°C |
Table 2. MS Conditions
|
Instrument |
Agilent 5975C MSD |
|
Source temp |
230°C |
|
Quadrupole temp |
150°C |
|
MSD mode |
SIM |
The monitored ions for each compound are listed in table 3 below. Ions were acquired using Selected Ion Monitoring (SIM) mode.
Table 3. Ions monitored for each analyte
|
Compound |
Quantifier Ion |
1st Qualifier Ion |
2nd Qualifier Ion |
|
Carbofuran 1 |
164 |
149 |
122 |
|
Propamocarb |
58 |
42 |
44 |
|
Mevinphos |
127 |
109 |
192 |
|
Acephate |
136 |
42 |
94 |
|
Captan |
79 |
80 |
77 |
|
Carbaryl 1 |
144 |
115 |
116 |
|
Methiocarb 1 |
168 |
153 |
109 |
|
Propoxur |
110 |
152 |
27 |
|
Ethoprophos |
158 |
43 |
97 |
|
Dimethoate |
87 |
93 |
125 |
|
Carbofuran 2 |
164 |
149 |
122 |
|
Atrazine |
200 |
215 |
58 |
|
Quintozene |
237 |
249 |
295 |
|
Pyrimethanil |
198 |
77 |
199 |
|
Diazinon |
137 |
179 |
152 |
|
Parathion-methyl |
263 |
125 |
109 |
|
Carbaryl 2 |
144 |
115 |
116 |
|
Metalaxyl |
45 |
132 |
206 |
|
Pirimiphos-methyl |
290 |
276 |
305 |
|
Methiocarb 2 |
168 |
153 |
109 |
|
Malathion |
173 |
125 |
93 |
|
Chlorpyrifos |
197 |
97 |
199 |
|
Oxamyl |
72 |
42 |
69 |
|
Cyprodinil |
224 |
77 |
225 |
|
Thiabendazole |
201 |
174 |
63 |
|
Fluopyram |
173 |
145 |
223 |
|
Fipronil |
367 |
369 |
213 |
|
Chlordane |
373 |
375 |
377 |
|
Prallethrin 1 |
123 |
81 |
77 |
|
Prallethrin 2 |
123 |
81 |
77 |
|
Imazalil |
215 |
41 |
173 |
|
Isoprothiolane |
118 |
162 |
189 |
|
Buprofezin |
105 |
106 |
172 |
|
Myclobutanil |
179 |
152 |
181 |
|
Fludioxonil |
248 |
127 |
154 |
|
Trifloxystrobin |
116 |
131 |
59 |
|
Cyflufenamid |
91 |
55 |
118 |
|
Chlorfenapyr |
59 |
60 |
247 |
|
Penthiopyrad |
177 |
302 |
152 |
|
Quinoxyfen |
237 |
272 |
307 |
|
Propiconazole |
173 |
69 |
259 |
|
DDT |
235 |
237 |
176 |
|
Kresoxim-methyl |
116 |
131 |
206 |
|
Fluopicolide |
209 |
210 |
211 |
|
Tebuconazole |
125 |
250 |
70 |
|
Propargite |
135 |
81 |
57 |
|
Piperonyl butoxide |
176 |
149 |
177 |
|
Iprodione |
56 |
316 |
43 |
|
Fenoxycarb |
116 |
88 |
255 |
|
Bifenthrin |
181 |
165 |
|
|
Fluxapyroxad |
159 |
160 |
|
|
Acetamiprid |
56 |
152 |
126 |
|
Fenpropathrin |
97 |
55 |
181 |
|
Etoxazole |
204 |
300 |
141 |
|
Fenamidone |
238 |
237 |
239 |
|
Cyflumetofen |
173 |
145 |
|
|
Pyriproxyfen |
136 |
78 |
77 |
|
Lambda-Cyhalothrin |
181 |
197 |
208 |
|
Ametoctradin |
176 |
190 |
204 |
|
Permethrin 1 |
183 |
41 |
27 |
|
Pyridaben |
147 |
117 |
148 |
|
Permethrin 2 |
183 |
41 |
27 |
|
Coumaphos |
362 |
97 |
109 |
|
Cyfluthrin 1 |
163 |
165 |
206 |
|
Fenbuconazole |
129 |
198 |
125 |
|
Cyfluthrin 2 |
163 |
165 |
206 |
|
Boscalid |
140 |
112 |
142 |
|
Cypermethrin 1 |
163 |
181 |
165 |
|
Cypermethrin 2 |
163 |
181 |
165 |
|
Cypermethrin 3 |
163 |
181 |
165 |
|
Etofenprox |
163 |
135 |
107 |
|
Difenoconazole |
265 |
323 |
267 |
|
Indoxacarb |
59 |
|
|
|
Azoxystrobin |
344 |
388 |
345 |
|
Famoxadone |
330 |
329 |
331 |
|
Dimethomorph |
301 |
165 |
303 |
Results
Using the automated cSPE procedure described in this applica- tion note, a panel of 77 pesticides spiked into oat extracts were cleaned-up using cSPE. All 77 pesticides demonstrated recovery between the acceptable limits of 70-130%, with the majority being just above 90%. Figure 2 shows recoveries obtained from oat extract spiked prior to cSPE clean-up at a concentration equivalent to 96 ppb in dry oat matrix. Excellent reproducibility was achieved, all 77 analytes demonstrating RSDs lower than 20% (n=4), with most being below 5%. Figure 3 shows %RSD obtained from oats extract spiked prior to cSPE clean-up at a concentration equivalent to 96 ppb in dry oat matrix. Automation of the method ensures variation is kept to a minimum. Figure 2. Average analyte % recoveries obtained from oat extract spiked at a concentration equivalent to 96 ppb in dry oat matrix (n=4).
Figure 2. Average analyte % recoveries obtained from oat extract spiked at a concentration equivalent to 96 ppb in dry oat matrix (n=4).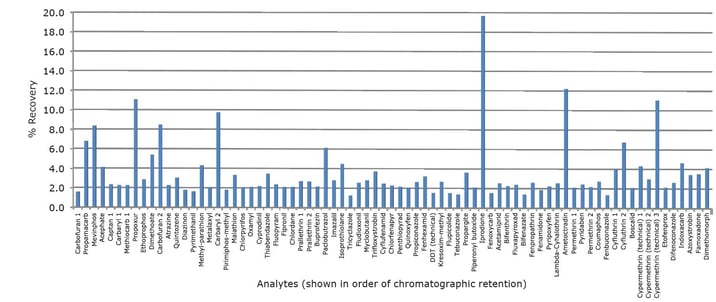
Figure 3. %RSDs obtained from oat extract spiked at a concentration equivalent to 96 ppb in dry oat matrix (n=4).
Linearity and LOQ
Calibration curves were produced for oat extract spiked with pesticides, either within the range 0.8 ppb – 160 ppb or the range 4 ppb to 800 ppb depending on analyte limit of quantitation (LOQ).
To ensure that calibration samples reflect real samples as closely as possible, samples were prepared using pooled oat extract (from QuEChERS step 1). The highest-level spiked sample was prepared at a concentration of 800 ppb. The remaining spiked samples were prepared by a serial dilution of the highest-level spiked sample using additional blank oat extract. 1 mL of each concentration of calibration samples were then cleaned-up using cSPE cartridges.
LOQs were estimated at the lowest concentration of which the analyte gave a signal to noise ratio of 10:1. LOQs of analytes varied across the large panel with most being between 0.8 and 4 ppb. Good linearity was observed for most analytes, typically giving r2 values greater than 0.99. Table 4 below shows the r2 and LOQ values of all analytes.
|
Compound |
r2 |
LOQ (ppb) |
Compound |
r2 |
LOQ(ppb) |
|
Carbofuran 1 |
0.999 |
0.8 |
Fenhexamid |
0.992 |
8* |
|
Propamocarb |
0.996 |
0.8 |
DDT |
0.999 |
0.8 |
|
Mevinphos |
0.999 |
4 |
Kresoxi-methyl |
0.998 |
0.8 |
|
Acephate |
0.999 |
40* |
Fluopicolide |
0.999 |
0.8 |
|
Captan |
0.992 |
4 |
Tebuconazole |
0.999 |
0.8 |
|
Carbaryl 1 |
0.996 |
4 |
Propargite |
0.997 |
0.8 |
|
Methiocarb |
0.999 |
0.8 |
Piperonyl butoxide |
0.999 |
0.8 |
|
Propoxur |
0.990 |
4 |
Iprodione |
0.999 |
4 |
|
Ethoprophos |
0.999 |
4 |
Fenoxycarb |
0.994 |
4 |
|
Dimethoate |
0.994 |
4 |
Acetamiprid |
0.995 |
8* |
|
Carbofuran 2 |
0.990 |
8 |
Bifenthrin |
0.999 |
0.8 |
|
Atrazine |
0.999 |
0.8 |
Fluxapyroxad |
0.999 |
0.8 |
|
Quintozene |
0.999 |
4 |
Bifenazate |
0.992 |
4 |
|
Diazinon |
0.996 |
4 |
Fenpropathrin |
0.998 |
8* |
|
Pyrimethanil |
0.999 |
0.8 |
Fenamidone |
0.990 |
4 |
|
Parathion-methyl |
0.999 |
4 |
Pyriproxyfen |
0.999 |
0.8 |
|
Metalaxyl |
0.997 |
4 |
Lambda- Cyhalothrin |
0.999 |
1.6 |
|
Carbaryl 2 |
0.996 |
8* |
|||
|
Pirimiphos-methyl |
0.999 |
0.8 |
Ametoctradin |
0.999 |
0.8 |
|
Malathion |
0.990 |
4 |
Permethrin 1 |
0.995 |
4 |
|
Chlorpyrifos |
0.996 |
4 |
Pyridaben |
0.995 |
4 |
|
Oxamyl |
0.998 |
40* |
Permethrin 2 |
0.995 |
4 |
|
Cyprodinil |
0.999 |
0.8 |
Coumaphos |
0.999 |
0.8 |
|
Thiabendazole |
0.994 |
4 |
Fenbuconazole |
|
0.999 |
|
Fluopyram |
0.995 |
4 |
Cyfluthrin 1 |
0.999 |
4 |
|
Fipronil |
0.999 |
0.8 |
Cyfluthrin 2 |
0.999 |
8* |
|
Chlordane |
0.999 |
0.8 |
Boscalid |
0.999 |
0.8 |
|
Prallethrin 1 |
0.999 |
8* |
Cypermethrin 1 |
0.999 |
8* |
|
Prallethrin 2 |
0.999 |
8* |
Cypermethrin 2 |
0.998 |
8* |
|
Paclobutrazol |
0.999 |
0.8 |
Cypermethrin 3 |
0.999 |
8* |
|
Imazalil |
0.991 |
4 |
Etofenprox |
0.999 |
0.8 |
|
Isoprothiolane |
0.999 |
0.8 |
Difenoconazole |
0.991 |
8 |
|
Tricyclazole |
0.993 |
4 |
Indoxacarb |
0.999 |
40* |
|
Fludioxonil |
0.9999 |
0.8 |
Azoxystrobin |
0.995 |
4 |
|
Buprofezin |
0.995 |
4 |
Famoxadone |
0.996 |
4 |
|
Myclobutanil |
0.994 |
4 |
Dimethomorph |
0.999 |
1.6 |
|
Trifloxystrobin |
0.999 |
0.8 |
|
|
|
|
Cyflufenamid |
0.995 |
8* |
*analytes have higher LOQs than most of the panel so have less than 6 calibration points |
||
|
Chlorfenapyr |
0.999 |
1.6 |
|||
|
Penthiopyrad |
0.999 |
0.8 |
|
||
|
Quinoxyfen |
0.999 |
0.8 |
|
||
|
Propiconazole |
0.995 |
4 |
|
||
Calibration curves
Calibration curves for selected pesticides are show in figures 4-7 below. Figure 4. Atrazine, calibration range 0.8 to 160 ppb
Figure 4. Atrazine, calibration range 0.8 to 160 ppb![]() Figure 5. Propiconazole, calibration range 4 to 800 ppb
Figure 5. Propiconazole, calibration range 4 to 800 ppb Figure 6. Trifloxystrobin, calibration range 0.8 to 160 ppb
Figure 6. Trifloxystrobin, calibration range 0.8 to 160 ppb Figure 7. Azoxystrobin, calibration range 4 to 800 ppb
Figure 7. Azoxystrobin, calibration range 4 to 800 ppb
Discussion and conclusion
The use of ISOLUTE® cSPE for QuEChERS cartridges provides a simple, robust flow-through clean-up of pesticides in QuEChERS extracts from oat samples for pesticide analysis. The cartridges deliver high recoveries, low RSDs and exceptionally clean extracts.
Excellent linearity was achieved across a wide calibration range for all analytes, demonstrating that the flow-through cSPE approach provides consistent analyte recovery and matrix removal at all concentration levels, with LOQ typically < 1 ppb.
When automated using a Biotage® Extrahera™ workstation, a batch of 24 samples can be cleaned-up in 10.3 minutes (15.1 minutes for a batch of 48 samples). ISOLUTE® cSPE for QuEChERS cartridges provide a simplified workflow for the clean-up step compared to the dSPE equivalent (see figure 8 page 7), which results in fewer manual transfer steps and higher throughput. The equivalent manual dSPE procedure takes approx. 23.5 minutes to clean-up 24 samples and ~47 minutes to clean-up 48 samples.
Traditionally QuEChERS using dSPE for clean-up does not utilize a concentration step. However, standard QuEChERS methods (AOAC, EN and mini-multiresidue) allow for an evaporative concentration step where large volume injection GC-MS analysis is not used, or where solvent exchange from acetonitrile is beneficial. Typically, a concentration factor of x 4 is utilized.
 Figure 8. Comparison of dSPE vs cSPE clean-up workflow
Figure 8. Comparison of dSPE vs cSPE clean-up workflow
In order to improve our working sample concentration range we incorporated a post clean-up evaporation and reconstitu¬tion step. After cSPE clean-up, the extract was evaporated to dryness under highly controlled conditions using the TurboVap® 96 Dual, and reconstituted in a total volume of 30 μL of toluene, from which 1.3 μL was injected. No significant evaporative losses were observed.
This represents a 30 x increase in analyte concentration injected into the GC-MS system, and allows for a more appropriate injec- tion solvent for GC-MS analysis. The higher concentration factor allows for improved sensitivity (lower analyte LOQ) using the single quadrupole GC-MS system. It is expected that LOQs could be further improved using a higher sensitivity / more selective GC-MS/MS system.
The improved cleanliness of the extract from the flow through cSPE process (more efficient matrix removal) compared with dSPE means that the more concentrated sample extract (x 30 compared with x 4) can be injected without compromising the GC system or analyte separation.
Chemicals and reagents
- All pesticide stock solutions were purchased from LGC Ltd. (Middlesex, UK) and stored at -20°C.
- California Pesticide Class 1, Class 2A, Class 2B mixes were purchased at 100 µg/mL. All other compounds were purchased as individual stocks at 1 mg/mL and diluted to 100 µg/mL with acetonitrile.
- A spiking solution was prepared weekly in acetonitrile at a concentration of 10 µg/mL and stored at -20°C.
- Acetonitrile was purchased from Rathburn Chemicals Ltd. (Walkerburn, UK).
Additional information
Preparation of pooled sample for cSPE evaluation
The focus of this application note is evaluation of the automated clean-up step using cSPE cartridges. Therefore, to provide homogenous samples for comparison purposes, supernatants prepared in step 1 were pooled and spiked appropriately with pesticide standards. 1 mL aliquots of the pooled supernatants were used for evaluation of the cSPE clean-up step.
For recovery testing, the pooled sample was spiked at a concentration of 120 ng/mL which equates to 96 ppb in dry oat matrix. This is calculated with the assumption that 10 g of oat matrix with QuEChERS salts results in 8 mL of supernatant for cSPE clean up. Supernatant volumes will depend on matrix type and initial QuEChERS salt used.
For determination of linearity and LOQ, the highest-level spiked sample was prepared at a concentration of 800 ppb. The remaining spiked samples were prepared by a serial dilution of the highest-level spiked sample with the pooled blank oat extract.
Use of internal standards
No internal standardisation was used in this application note, however if being used, we would recommend that appropriate internal standards were added to the homogenised matrix along with acetonitrile and QuEChERS salts during step 1, prior to mixing and subsequent centrifugation.
Evaporation and reconstitution
The use of the tapered bottom microsampling vials (Agilent p/n 5184-3550) for extract collection, evaporation and reconstitution reduces variability in this step.
Ordering information
|
Biotage® Consumables Ordering Information |
||
|
Part Number |
Description |
Pack Size |
|
Homogenization and QuEChERS extraction used in this application note |
||
|
Q0020-15V |
Extraction salts: ISOLUTE® QuEChERS EN 10 g/15 mL Extraction Tube |
25 |
|
19-6650 |
Bulk 50 mL Tubes with Leak Proof Screw Caps |
100 |
|
19-640 |
Bulk 2.4 mm Metal Beads, 500 g |
1 |
|
QuEChERS Clean-up used in this application note |
||
|
Q0035-0020-BG |
cSPE: ISOLUTE® EN General 200 mg/3 mL (Tabless) |
50 |
|
414141 |
1000 µL Clear Tips |
960 |
|
Biotage ® Equipment & Accessories Ordering Information |
||
|
Part Number |
Description |
Pack Size |
|
Homogenization and QuEChERS extraction |
||
|
19-060 |
Biotage® Lysera |
1 |
|
19-345-050 |
50 mL Tube Carriage Kit |
1 |
|
QuEChERS Clean-up |
||
|
414001 |
Biotage® Extrahera™ Classic |
1 |
|
417002 |
Biotage® Extrahera™ HV-5000 |
1 |
|
414174SP |
Column Rack 24 x 3 mL |
1 |
|
415555SP |
Sample/Collection Rack 12 x 75 mm, 48 Positions |
1 |
|
414511SP |
Collection Rack 12 x 75 mm, 24 Positions |
1 |
|
414578SP |
Inserts For 12 x 32 mm Vials For Collection Rack 12 x 75 mm 24 positions |
24 |
|
414256SP |
Sample Rack 12 x 75 mm, 24 Positions |
1 |
|
Evaporation |
||
|
418000 |
TurboVap® 96 Dual |
1 |
|
418319SP |
Rack 12 x 32 mm, 24 Positions |
1 |
|
ISOLUTE® cSPE for QuEChERS Range |
||
|
Selection of the appropriate product depends on regulation (AOAC or EN) and matrix type. |
||
|
Part Number |
Description |
Pack Size |
|
QuEChERS extraction |
||
|
Q0010-15V |
Extraction salts: ISOLUTE® QuEChERS AOAC 15 g/15 mL Extraction Tube |
25 |
|
Q0020-15V |
Extraction salts: ISOLUTE® QuEChERS EN 10 g/15 mL Extraction Tube |
25 |
|
QuEChERS Clean-up |
||
|
Q0030-0020-BG |
ISOLUTE® AOAC General 200 mg/3 mL (Tabless) |
50 |
|
Q0035-0020-BG |
ISOLUTE® EN General 200 mg/3 mL (Tabless) |
50 |
|
Q0050-0035-BG |
ISOLUTE® AOAC Waxed 350 mg/3 mL (Tabless) |
50 |
|
Q0060-0035-BG |
ISOLUTE® EN Waxed 350 mg/3 mL (Tabless) |
50 |
|
Q0070-0015-BG |
ISOLUTE® AOAC Pigment 150 mg/ 3 mL (Tabless) |
50 |
|
Q0080-0030-BG |
ISOLUTE® EN Pigment 300 mg/3 mL (Tabless) |
50 |
|
Q0090-0050-BG |
ISOLUTE® EN High Pigment 500 mg/3 mL (Tabless) |
50 |
Appendix 1: Biotage® Extrahera™ classic method
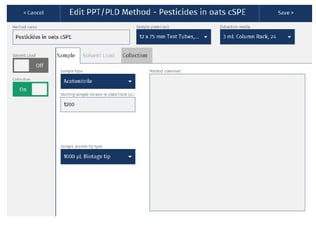
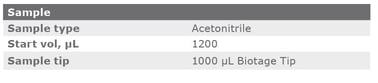
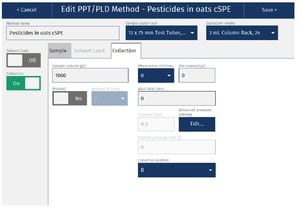
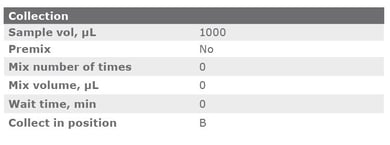
The cSPE clean-up method described in this application note was automated on the Biotage® Extrahera™ Classic. This appendix contains the software settings required to configure Extrahera™ Classic to run this method for a 1 mL sample aliquot.
|
Sample plate/rack |
12 x 75 mm, 24 |
|
Extraction media |
1 mL Column Rack, 24 (ISOLUTE® EN General) |
|
Sample Load |
Off |
|
Collection |
On |

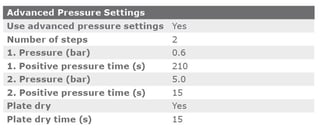
Appendix 2: Options for streamlining the automated QuEChERS process
Homogenization & Extraction
As part of the QuEChERS workflow, use of the Biotage® Lysera for initial sample pre-treatment and extraction (QuEChERS step 1) reduces the need for manual steps, saving time and reducing the possibility of manual handling errors. The need to clean a blending device in-between each sample is eliminated and an even homogenisation between each sample is ensured.
In this application note, ISOLUTE® EN QuEChERS extraction salts (p/n Q0020-15V) optimized for extraction of 10 g of sample matrix were used to prepare samples in 50 mL tubes, for the initial extraction step.
In place of manually shaking samples by hand, 3 x 50 mL tubes can be shaken simultaneously in 15 seconds using the Biotage Lysera.
Clean-up
Using the Biotage® Extrahera™ HV-5000 platform for automated clean-up (instead of the Extrahera™ Classic platform used for this application note), 50 mL extraction tubes from the initial homogenization and extraction step can be transferred directly to the Extrahera™ HV-5000 after centrifugation, eliminating a manual sample transfer step. The supernatant can then be aspirated directly from the 50 mL centrifuge tube to the cSPE cartridge utilizing the Extrahera’s ‘Smart Pipetting’ function, ensuring no particulate is transferred. A maximum of 12 samples can be processed using 50 mL tubes.
Increasing batch size and sample throughput with minimal manual sample transfer
As an alternative to 50 mL tubes for homogenization and extraction, Biotage® Lysera can be used to homogenize samples in smaller tubes (30 mL, 7 mL) simultaneously processing batch sizes of up to 12 samples.
Reduction of the starting mass of sample, for example from 15 g to 5 g or 2 g, extracted with an appropriately reduced wight of extraction salt, and utilizing smaller extraction tubes can lead to increased batch size and further reduction in overall processing time.
After centrifuging these can be transferred directly to the Extrahera™ for cSPE clean-up, with no additional manual sample transfer steps. These options are summarized in the table below.
Options for increasing batch size with minimal manual sample transfer
|
Matrix sample size |
Extraction tube volume |
Biotage® Lysera capacity |
Maximum batch size using Biotage® Extrahera™ HV-5000 |
|
10-15 g |
50 mL |
3 tubes |
12 |
|
5-10 g |
30 mL |
6 tubes |
12 |
|
1.5-2.5 g |
7 mL |
12 tubes |
24 |
Alternatively, a manual transfer of the supernatant into test tubes can be included after QuEChERS extraction, which can then be processed in a batch size of up to 48.
Manual processing options
Clean-up of pre-extracted samples using ISOLUTE® cSPE for QuEChERS cartridges can be performed manually using the Biotage® Pressure+ 48 Positive Pressure Manifold or Biotage® VacMaster™-20. Processing parameters are available on request.
Literature Number: AN1002

