Synthesis of a branched peptidoglycan mimic and Multiple Antigenic Peptide (MAP) using Branches™
By Biotage
A branched peptidoglycan mimic and a tetra-branched antimicrobial peptide analogue
were synthesized on a lysine scaffold using Biotage® Initiator+ Alstra™ microwave peptide synthesizer. These peptide modifications are challenging to synthesize and automate, however, the procedure was operationally simplified using Branches™.
Introduction
Branched peptides are of great interest due to their therapeutic potential.1 Many interesting peptides have a branched structure such as multiple antigenic peptides (MAP)2 and peptidoglycans.3 Synthesis of branched peptides can be very challenging and automating such a synthesis is also difficult. Here we demonstrate the synthesis of a branched peptidoglycan mimic 1 and a tetra-branched antimicrobial peptide analogue 2 synthesized on a lysine scaffold and programmed using Branches™ to overcome the synthesis automation challenges.
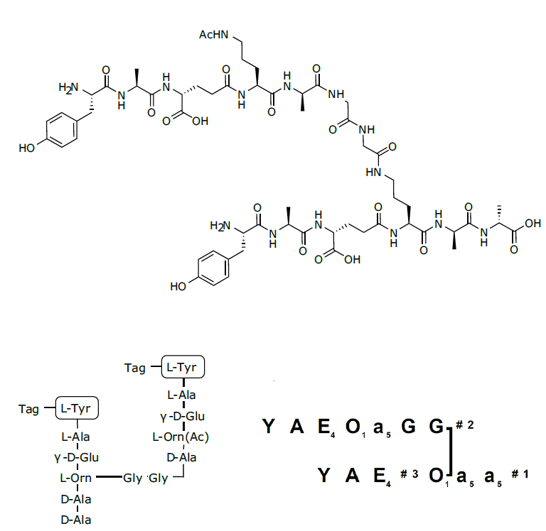
Figure 1: Branched peptidoglycan mimic 1 and Branches™ representation of 1.
Peptidoglycans are central structural components of the cell wall of bacteria. Several plant receptors are known to recognize peptidoglycan fragments. It is believed that these receptors form part of the defence mechanism against bacterial infections in several plant species. Synthesizing fragments or analogues of the structures is useful when investigating the biosynthesis function and recognition of peptidoglycans. The peptidoglycan fragment 1 from T. thermophilus contained an Orn(Gly-Gly) cross-link between the peptide strands and was synthesized as a putative substrate of T. thermophilus P60, a D,L-endo- peptidase involved in peptidoglycan re-modelling during bacterial growth.4 Furthermore, the multiple antigenic peptide (MAP) 2 synthesized was based on the well-known 10-mer antimicrobial peptide anoplin found in the venom of the solitary wasp Anoplius samariensis.5
Figure 2: Tetra-branched antimicrobial peptide analogue (MAP) 2 and Branches™ representation of 2.
Experimental
Materials
All materials were obtained from commercial suppliers; Sigma- Aldrich (acetonitrile, formic acid, triethylsilane (TES), hydrazine and dichloromethane (DCM)), Iris Biotech GmbH (Fmoc-amino acids, N,N-dimethylformamide (DMF), N-methylpyrrolidone (NMP), N-[(1H-benzotriazol-1-yl)(dimethylamino)methylene]-N- methylmethanaminium hexafluorophosphate N-oxide (HBTU), 1-Hydroxy-7-azabenzotriazole (HOAt), trifluoroacetic acid (TFA), piperidine and N,N-diisopropylethylamine (DIPEA)) and Rapp Polymere GmbH (TentaGel S Trt Cl resin and TentaGel R Rink Amide resin). Milli-Q (Millipore) water was used for LC-MS analysis.
Nα-9-fluorenylmethoxycarbonyl (Fmoc) amino acids contained the following side-chain protecting groups: tert-butyl (Tyr, Thr), 2,2,4,6,7-pentamethyl-dihydrobenzofuran-5-sulfonyl (Pbf, for Arg) and tert-butoxycarbonyl (Boc, for Lys) and 1-(4,4-dimethyl- 2,6-dioxocyclohex-1-ylidene)ethyl (Dde, for Lys), α-benzyl ester (α-OBn, for Glu). Ornithine (Orn) was Nα-protected with 1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)ethyl (Dde) and the side chain protecting group was Fmoc. Fmoc-GluOBn was made in-house using a novel microwave heating method and will be described elsewhere.
Synthesis
General solid-phase synthesis of peptides
All peptides were prepared by Fmoc solid-phase peptide synthesis on a Biotage® Initiator+ Alstra™ fully automated microwave peptide synthesizer. The synthesis of 1 was carried out on TentaGel S Trt Cl resin and the synthesis of 2 was carried out on TentaGel R Rink Amide resin.
Fmoc deprotections were performed at room temperature in two stages with the first using piperidine-DMF (2:3) for 3 min, followed by piperidine-DMF (1:4) for 15 minutes. For the synthesis of the MAP peptide 2, one additional deprotection step was performed using piperidine-DMF (1:4) for 15 minutes. Peptide couplings for the linear peptides were performed using 5.2 eq. of amino acid, 5.2 eq. of (HOAt), 5.0 eq. of HBTU all dissolved in DMF, and 9.38 eq. of DIPEA in NMP for peptide 1 and 16 eq. of amino acid, 16 eq. of HOAt, 15.2 eq. of HBTU all dissolved in DMF, and 28.8 eq. of DIPEA in NMP for peptide 2 where four peptide branches are synthesized simultaneously. A coupling time of 10 minutes at 75 °C was employed. After each coupling and deprotection step, a washing procedure with NMP (x 4), DCM (x 1) and then DMF (x 1) was employed.
After the synthesis was completed the resin was washed with DCM (x 5) and thoroughly dried. Peptides were cleaved from the resin with TFA/H2O/TES (95:3:2) for 5 min. and then for 2 h. The peptides were precipitated with cold diethyl ether. Analysis of the peptides was performed by LCMS on a Dionex Ultimate 3000 with Chromeleon 6.80SP3 software coupled to an ESI-MS (MSQ Plus Mass Spectrometer, Thermo). The peptides were analyzed on a Biotage® Resolux™ 300 Å C18 column (5 μm, 150 × 4.6 mm) with a flow rate of 1.0 mL/min. The following solvent system was used: solvent A, water containing 0.1% formic acid; solvent B, acetonitrile containing 0.1% formic acid. The column was eluted using a linear gradient from 5%–100% of solvent B.
Results & Discussion
Synthesis of peptidoglycan mimic 1
H-L-Tyr-L-Ala-γ-D-Glu-L-[Nγ-(H-L-Tyr-L-Ala-γ-D-Glu-L-Orn(Nγ-Ac)-D-Ala-L-Gly-L-Gly-)]Orn-D-Ala-D-Ala-OH
The branched peptide sequence 1 was assembled using SPPS as described above (Scheme 1) using Dde-Orn(Fmoc)-OH at the branching points. The N-terminal tyrosines were included as a tag for aiding HPLC analysis. The synthesis was carried out on TentaGel S Trt Cl resin (loading 0.25 g/mmol) on a 0.05 mmol scale in a 5 mL reactor vial. Loading of the first amino acid was achieved using 10 eq. Fmoc-D-Ala-OH dissolved in dry DMF together with 5 eq. dry DIPEA in dry NMP which was added to the resin and reacted for 16 hours and the resin washed with NMP (x 4), DCM (x 1) and then DMF (x 3).
Note: this quantity of resin is over the recommended scale range but was achievable by reducing the working volume (HOAt was added as a solid directly to the Fmoc-amino acid rather than as a separate solution).
Fmoc-D-Ala-OH and Dde-L-Orn(Fmoc)-OH were subsequently coupled using the method described above. After removal of the Fmoc moiety from the side-chain of the ornithine residue, the branch was extended by coupling on the δ-amino group with Fmoc-L-Gly-OH, Fmoc-L-Gly-OH, Fmoc-D-Ala-OH, and Dde-L-Orn(Fmoc)-OH. The Fmoc group from the side-chain of the second Orn was removed, the γ-amino group was acetylated by treatment with 20% acetic anhydride in DMF for 15 min (x2) and then washed with NMP (x 4), DCM (x 3) and then DMF (x 3). The two Nα-Dde groups of Orn were then removed by treatment with 1% hydrazine hydrate in DMF for 250 min (x 2) at room temperature and then washed thoroughly with NMP (x 4), DCM (x 3) and DMF (x 3).
Fmoc-D-Glu-OBn was subsequently coupled through the glutamic acid γ-side-chains, followed by Fmoc-L-Ala-OH, and Fmoc-L-Tyr-OH. After Fmoc deprotection of the two N-terminal amino groups, the peptide was cleaved from the resin as described above. Finally, the two α-benzyl ester groups of Glu were removed by treatment with hydrogen in the presence of 10% palladium on charcoal in methanol to afford the fully unprotected peptide 1, with a crude purity of 80% and confirmed by ESI-MS (Figure 3), calculated average isotopic composition for C59H86N14O22: 1341.6 Da. Found: m/z:1342.9 [M+H]+, 672.2 [M+2H]2+.
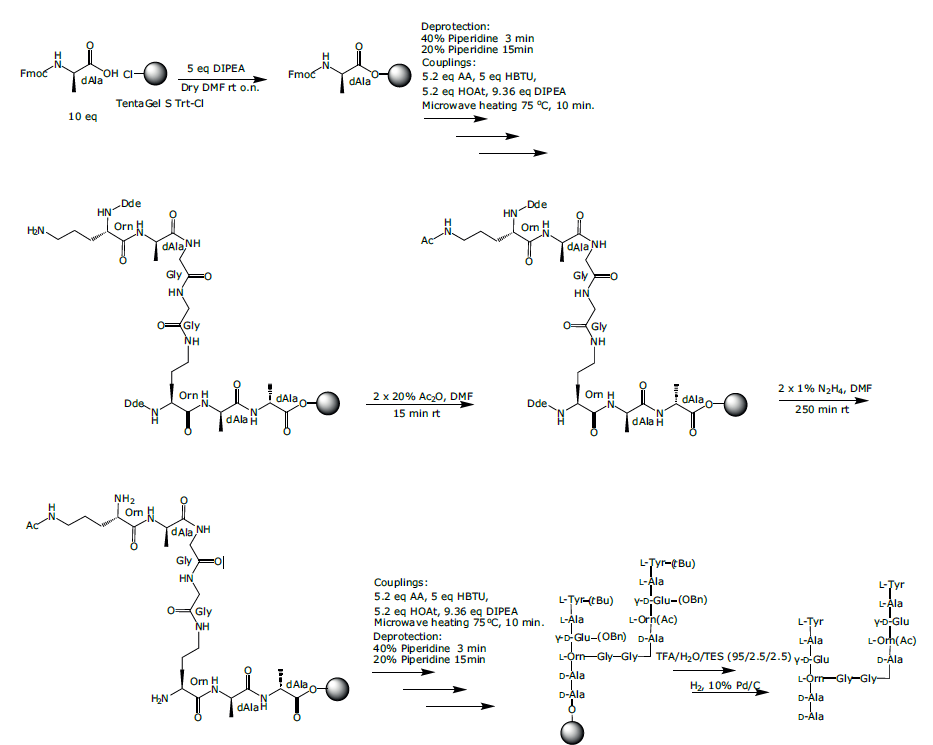
Scheme 1: Synthesis of branched peptidoglycan mimic 1.
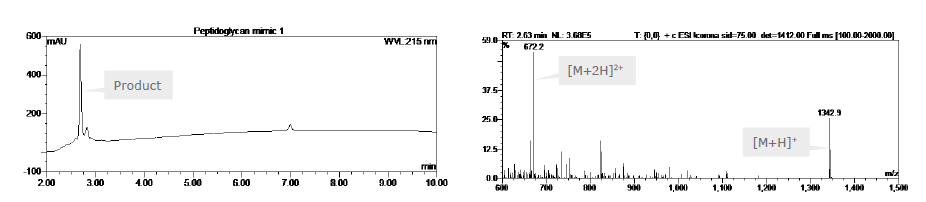
Figure 3: RP-HPLC chromatogram and ESI-MS of peptidoglycan mimic 1
Synthesis of the tetra-branched antimicrobial peptide analogue 2
The MAP peptide sequence 2 was assembled using SPPS as described above (Scheme 2) on a lysine scaffold. The synthesis was carried out on TentaGel R Rink resin (loading 0.18 g/mmol) on a 0.08 mmol scale in a 10 mL reactor vial. The first Fmoc-Lys(Fmoc)-OH was anchored to the resin using 4 eq. of Fmoc-Lys(Fmoc)-OH, 3.8 eq. HBTU, 4 eq. HOAt, 7.2 eq. DIPEA and reacted for 10 min. at 75 °C. The resin was washed NMP (x 3), DCM (x 1) and DMF (x 1). The Fmoc groups were removed with piperidine-DMF (2:3) for 3 min, followed by piperidine-DMF (1:4) for 15 minutes and the resin was then washed with NMP (x 4), DCM (x 1) and DMF (x 1).
This synthesis has some unique challenges in terms of programming the variable reagent quantities required at the beginning of the synthesis when constructing the lys scaffold compared to the simultaneous peptide branches synthesis. Initially we reduced the equivalents for the first two couplings by lowering the equivalents factor (in the software) to match the intended equivalents of amino acid coupling reagent and base. The reagent quantities for each cycle can be easily adjusted and customized in this way. Fmoc-Lys(Fmoc)-OH was coupled to the resin to create the first two branching points (equivalents factor changed from 1 to 0.25) using 4 eq. Fmoc-Lys(Fmoc)-OH, 3.8 eq. HBTU, 4 eq. HOAt, 7.2 eq. DIPEA, then washed and Fmoc deprotected as described above.
Fmoc-Lys(Fmoc)-OH was coupled to the the first two branching points (equivalents factor changed from 1 to 0.5) using 8 eq. Fmoc-Lys(Fmoc)-OH, 7.6 eq. HBTU, 8 eq. HOAt, 14.4 eq. DIPEA, then washed and Fmoc deprotected as described above. This created the four branching points required. The four branches were then elaborated with the 10-mer anoplin sequence on each branch as described above. Finally the four N-terminal Fmoc groups were removed, the resin washed then the peptide was cleaved as described above to afford the MAP peptide 2 and confirmed by ESI-MS (Figure 4), calculated average isotopic composition for C234H443N67O47: 4947.44 Da. Found: m/z: 1650.3 [M+3H]3+, 1237.9 [M+4H]4+,990.5 [M+5H]5+, 825.6 [M+6H]6+.
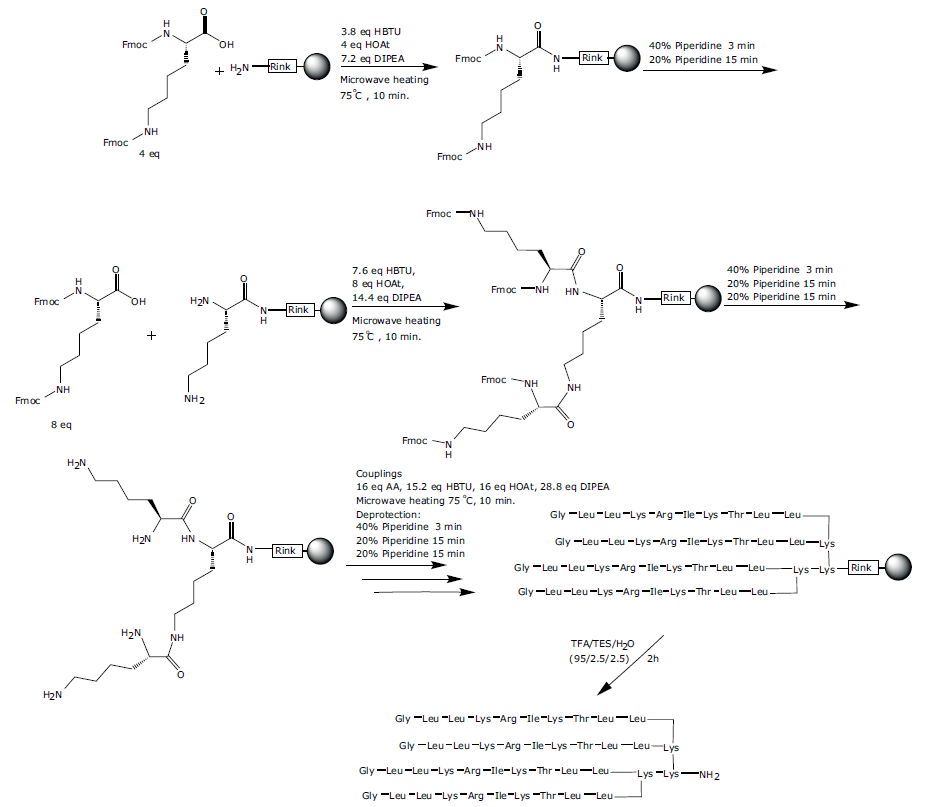
Scheme 2: Synthesis of the tetra-branched antimicrobial peptide analogue 2
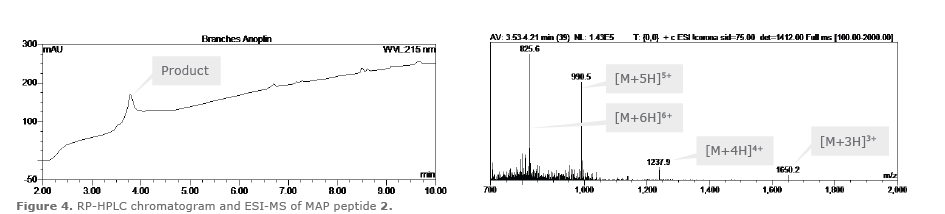
Figure 4: RP-HPLC chromatogram and ESI-MS of MAP peptide 2.
Conclusion
Here we have demonstrated the synthesis of a branched peptidoglycan mimic 1 and the synthesis of a tetra-branched antimicrobial peptide analogue (MAP) 2 on a lysine scaffold using Biotage® Initiator+ Alstra™ microwave peptide synthesizer. These peptide modifications are challenging to synthesize and automate, however, the procedure was operationally simplified using Branches™.
References
- Pini, A.; Falciani, C; Bracci, L., Curr. Protein Pept. Sci. 2009, 10, 194–5.
- Tam, J. P., Proc. Natl. Acad. Sci. USA 1988, 85, 5409–5413.
- Typas, A.; Banzhaf, M.; Gross, C. A.; Vollmer, W., Nat. Rev. Microbiol. 2012, 10, 123–136.
- Sørensen, K. K.; Thygesen, M. B.; Jensen, K. J. A New Approach to the Synthesis of Peptidoglycan Fragments. Presented at the 33rd European Peptide Symposium, Sofia, Bulgaria, August 31-September 5, 2014; Poster P068.
- Konno, K.; Hisada, M.; Fontana, R.; Lorenzi, C. C. B.;Naoki, H.; Itagaki, Y.; Miwa, A.; Kawai, N.; Nakata, Y.; Yasuhara, T.; Neto, J. R.; de Azevedo Jr, W. F.; Palma, M. S.; Nakajima, T., Biochimica et Biophysica Acta 2001, 1550, 70–80.
Acknowledgements
Prof. Knud J. Jensen and Dr. Kasper Kildegaard Sørensen at the University of Copenhagen.
Literature Number: AN097

