Automated synthesis of cyclic peptides on Biotage® Initiator+ Alstra™
By Biotage
Introduction
The interest in naturally occurring and designed bioactive cyclic peptides is ever increasing.1–4 Cyclic peptides often have an increased stability against proteolysis compared with linear peptides, which can be used in the design of potential peptide drugs. The decreased flexibility can provide more specific interactions with receptors. The fast and convenient synthesis of cyclic peptides is thus very appealing. The cyclic peptide mupain-1 (CPAYSRYLDC) is an inhibitor of the cancer-related urokinase-type plasminogen activator.5 Here we present two methods to assemble cyclic peptide analogues of mupain-1:
1. Cyclization via a disulphide bridge to give cyclic peptide 1
(Figure 1)
2. Side-chain to side-chain cyclization to give cyclic peptide 2
(Figure 2)
On-resin cyclization of peptides is very useful and now with Branches™, this can now be operationally simplified and save time.
Branches™ is a tool that allows skilled peptide chemists to assign custom methods and enables scheduling and visualization of operations, in order to make complex peptide modifications, using either fully automated or semi-automated protocols.
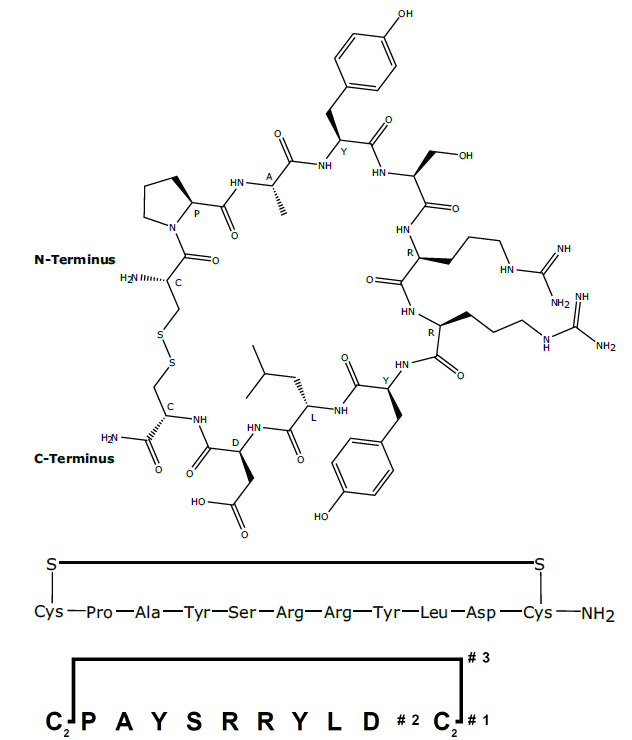
Figure 1: Cyclic peptide 1 and Branches™ representation of 1
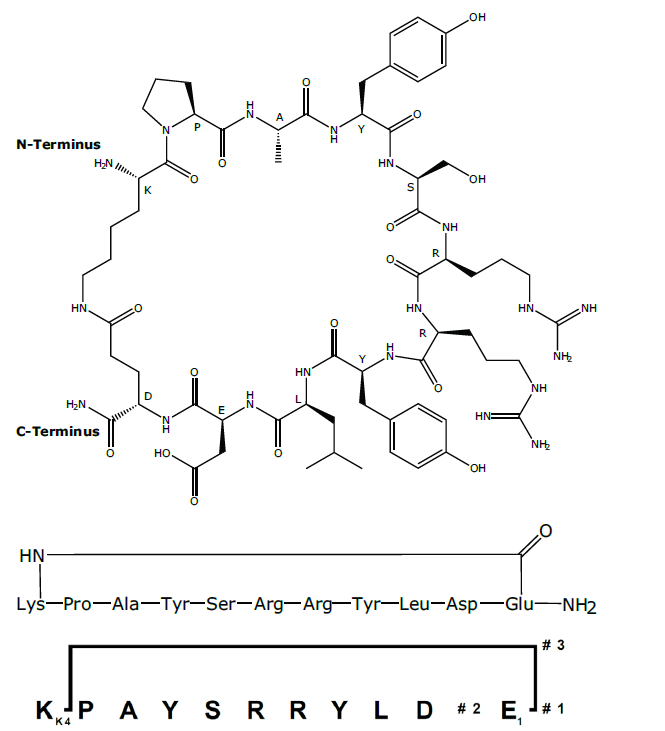
Figure 2: Cyclic peptide 2 and Branches™ representation of 2
Experimental
Materials
All materials were obtained from commercial suppliers; Sigma-Aldrich (acetonitrile, formic acid, borane dimethylamine complex, ammonia in methanol, dimethyl sulfoxide, hydrogen peroxide, tetrakis (triphenylphosphine)palladium(0)and triethyl silane (TES)), Iris Biotech GmbH (Fmoc-amino acids, N,N-dimethylformamide (DMF), N-methyl pyrrolidone (NMP), N-[(1H-benzotriazol-1-yl)(dimethyl amino)methylene]-N-methylmethanaminium hexafluorophosphate N-oxide(HBTU), 1-Hydroxy-7-azabenzotriazole (HOAt), trifluoroaceticacid (TFA), piperidine and N,N-diisopropylethylamine(DIPEA)) and Rapp Polymer GmbH (Tenta Gel S Rink Amideresin). Milli-Q (Millipore) water was used for LC-MS analysis. Nα-9-fluorenylmethoxycarbonyl (Fmoc) amino acids contained the following side-chain protecting groups: tert-butyl (Asp, Tyr,Ser), 2,2,4,6,7-pentamethyl-dihydrobenzofuran-5-sulfonyl (Pbf, for Arg), 4-methoxytrityl (Mmt, for Cys), allyloxycarbonyl (Alloc, for Lys) and γ-allyl ester (OAll, for Glu).
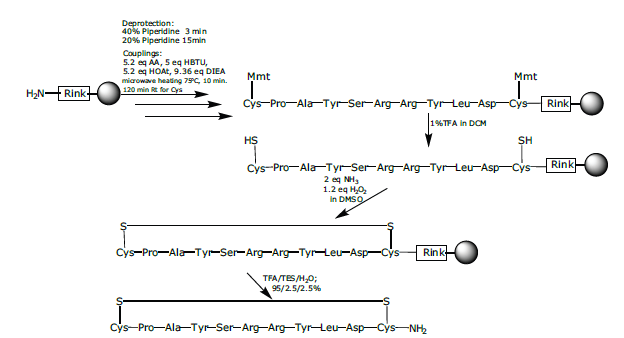
Figure 3: Synthesis of cyclic peptide 1.
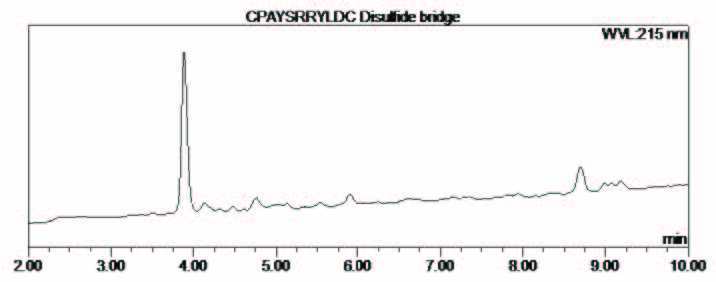
%20RP-HPLC-chromatogram-ESI-MS-synthesis-cyclic-peptide.jpg?width=719&height=281&name=biotage-an093-fgure4-(Right)%20RP-HPLC-chromatogram-ESI-MS-synthesis-cyclic-peptide.jpg)
Figure 4: (Right) RP-HPLC chromatogram and ESI-MS of the synthesis of cyclic peptide 1.
Synthesis
General solid-phase synthesis of peptides
All peptides were prepared by Fmoc solid-phase peptide synthesis on a Biotage® Initiator+ Alstra™ fully automated microwave peptide synthesizer. The syntheses were carried out on Tenta Gel S Rink Amide resin (0.25 mmol/g) on 0.11 mmol scale using a 10 mL reactor vial. Fmoc deprotections were performed at room temperature in two stages with the first using piperidine-DMF (2:3) for 3 min, followed by piperidine-DMF (1:4) for 10 minutes. Peptide couplings for the linear peptides were performed using 5.2 eq. of amino acid, 5 eq. of HOAt, 5 eq. of HBTU, and 9.36 eq. of DIPEA in NMP. A coupling time of 10 minutes at 75 °C was employed, After each coupling and deprotection steps, a washing procedure with NMP (x 3), DCM (x 1) and then DMF (x1) was employed. After the synthesis was completed the resin was washed with DCM (x 4) and thoroughly dried. Peptides were cleaved from the resin with TFA-H2O-TES (95:3:2) for 5min. and then for 2 h. The peptide was precipitated with cold diethyl ether. Analysis of the peptide was performed by LCMSon a Dionex Ultimate 3000 with Chromeleon 6.80SP3 software coupled to an ESI-MS (MSQ Plus Mass Spectrometer, Thermo).The peptide was analyzed on a Biotage®Resolux™300 Å C18column (5 μm, 150 × 4.6 mm) with a flow rate of 1.0 mL/min. The following solvent system was used: solvent A, water containing0.1% formic acid; solvent B, acetonitrile containing 0.1% formic acid. The column was eluted using a linear gradient from5%–100% of solvent B.
Results & discussion
Synthesis of cyclic peptide 1;
on-resin disulphide bridge cyclization
The linear peptide sequence was assembled using SPPS as described above with microwave heating during the coupling steps (Scheme 1). The Cys thiol moieties were protected by the very acid-labile protecting group 4-methoxytrityl (Mmt). The mild and selective removal of the Mmt groups was achieved by treatment with a solution of 1% TFA and 5% TES in DCM and reacted for 1 h. at room temperature followed by washing with DCM (x 1), DMF (x 8) and again with DCM (x 3). The extra DMF washes were carried out to remove any traces of TFA remaining in the fluid lines of the system. The peptidyl-resin was then treated with a dilute solution of ammonia and hydrogen peroxide in DMSO (2 eq. NH3 in methanol/1.2 eq. of H2O2 in DMSO) to establish the disulphide bridge. After 30 minutes the peptidyl-resin was washed with NMP (x 5), DCM (x 5) and then cleaved as described above to afford the desired cyclic peptide 1, with a crude purity of 83% and confirmed by ESI-MS (Figure 3), calculated average isotopic composition for C57H86N18O16S2, 1342.6 Da. Found: m/z 1343.8 [M+H]+, 671.7 [M+2H]2+. The software supported the fully automated synthesis of the cyclic peptide.
Synthesis of cyclic peptide 2;
side-chain to side-chain cyclization
The linear peptide sequence was assembled using SPPS as described above with microwave heating during the coupling steps (Scheme 2). After the linear synthesis was completed, the peptidyl-resin was washed with DCM (x 4). The Lys side chain was protected with an Alloc group while the Glu residue possessed a γ-allyl ester moiety. Following peptide chain assembly, the synthesis was pre-programmed with a pause step in order to facilitate the removal of both allyl protecting groups with Pd(O). This was achieved by treating the reaction with borane dimethylamine complex (Me2NH·BH3) (2.0 eq., 13 mg) in DCM (4 mL), degassing with argon using an argon balloon (30 min.), followed by the addition of tetrakis (triphenylphosphine)palladium(0) (Pd(PPh3)4), (1.0 eq., 127 mg) and allowing the reaction to proceed for 1 h. under continual degassing with argon. The resin was washed with DCM (x 4), DMF (x 4) and then the side-chain to side-chain cyclization (amide bond formation) was accomplished with HOAt (5.2 eq., 88 mg), HBTU (5 eq., 209 mg), and DIPEA (9.36 eq., 176 μL) with a coupling time of 10 min. at 75 °C.
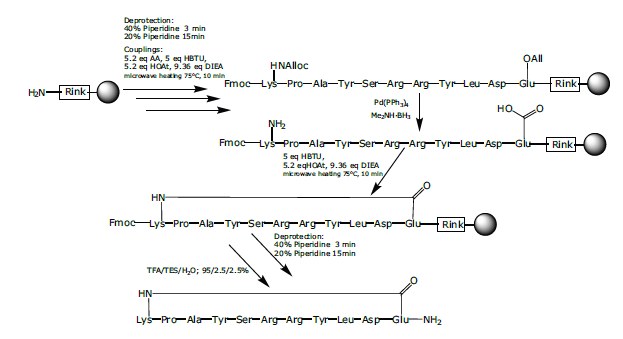
Figure 5: Synthesis of cyclic peptide 2.
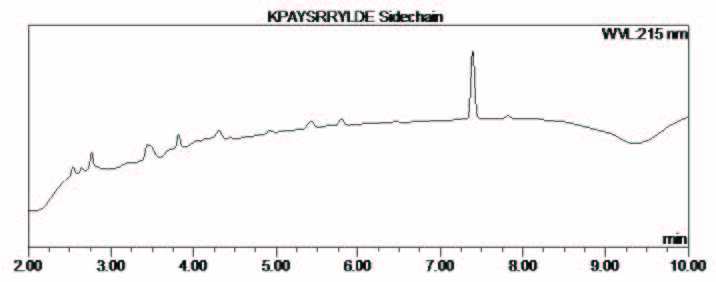
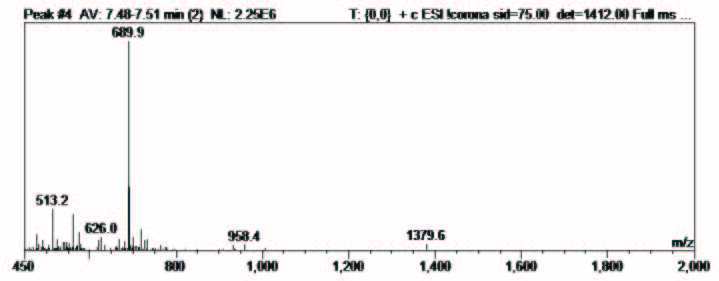
Figure 6: RP-HPLC chromatogram and ESI-MS of the synthesis of cyclic peptide 2.
The resin was washed with NMP (x 3), DCM (x 1), DMF (x 1) and then the N-terminal Fmoc group was removed with piperidine- DMF (2:3) for 3 min, followed by piperidine-DMF (1:4) for 10 minutes. The resin was washed and the peptide released from the solid support as described above to afford the desired cyclic peptide 2 with a crude purity of 85 % and confirmed by ESI-MS (Figure 4), calculated average isotopic composition for C62H95N19O17: 1378.56 Da. Found: m/z 1379.6 [M+H]+, 689.9 [M+2H]2+.
Conclusion
We have demonstrated the capability of the Biotage® Initiator+ Alstra™ microwave peptide synthesizer to fully automate the on-resin synthesis of cyclic peptides 1 and 2 with examples showing disulphide bridge formation and side-chain to side-chain cyclization respectively. The synthesis included specialized reactions of non-linear peptides which were achieved with a high degree of purity. Furthermore, this was achieved using the unique Branches™ tool which provides an extensive overview for the scheduling and visualization of operations in order to simplify the programming of complex peptide modifications and is a great addition to the toolbox for the peptide chemist.
References
1. Davies, J. S. J. Peptide Sci. 2003, 9, 471–501.
2. Panciera, M; Amorin, M; Granja, J. R. Chem. Eur. J. 2014, 20, 10260–10265.
3. Zheng, L; Marcozzi, A; Gerasimov, J. Y.; Herrmann, A. Angew. Chem. Int. Ed. 2014, 53, 7599–7603.
4. Scheinpflug, K; Nikolenko, H; Komarov, I. V; Rautenbach, M; Dathe, M. Pharmaceuticals. 2013, 6, 1130–1144.
5. Andersen, L. M; Wind, T; Hansen, H. D; Andreasen, P. A. Biochem. J. 2008, 412, 447–457.
Acknowledgements
Prof. Knud J. Jensen and Dr. Kasper Kildegaard Sørensen at the University of Copenhagen.
Literature Number: AN093

