Dec 20, 2023 10:12:06 PM
Does peptide length and size affect cyclization efficiency?
By Austin Schlirf

The big question…what affects cyclization efficiency? This is what I have been trying to answer over the course of the last few experiments, especially when it comes to head-to-tail macrocycles. I have investigated a strategy for cyclizing on resin and looked at loading capacity in my journey to figure out a starting point for peptide cyclization. In this blog I discuss how the length of peptide affects the efficiency of cyclization on resin.
Through my previous cyclization experiments I have used a peptide that is 15 amino acid residues long linked to my resin using the side chain of glutamic acid. My reactions thus far have yielded similar results typically seeing cyclization efficiency <30% with an equivalent amount of dimerization occurring. I have decreased the loading capacity to reduce dimerization, however this did not seem enough to improve the efficiency of this reaction nor reduce the formation of dimers. On top of that dimerization is really difficult to resolve chromatographically thus far and reducing this side product would just make life easier for me and ideally for you.
I assumed that the peptide chain length could promote dimerization and if I shortened this peptide, I would decrease dimerization by reducing chain-to-chain interaction. In this blog I test a 10-mer and 5-mer using my standard process for cyclization as described in my previous blogs. Ideally, the shorter peptides have less dynamic reach from one peptide chain to a different peptide chain limiting this dimerization. These sequences were chosen based on my original 15-mer maintaining the larger bulky groups at the N-terminus where applicable (Figure 1).
For one, my linear peptides were much purer compared to that of the 15-mer which is to be expected (Figure 1). This was promising as I would have less impurities convoluting my final cyclization. Remind me why I didn’t start with these?
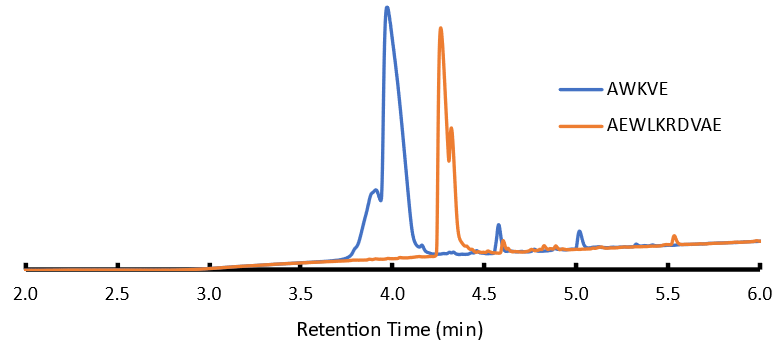
Figure 1: Chromatogram of linear, fully deprotected 10-mer and 5-mer. Peak splitting most likely due to 50/50 loading solvent mixture of ACN/H2O; MS showed no other significant species.
After removing the allyl protecting group from my C-terminal glutamic acid on the linear peptides, I once again cyclized my peptide using DIC/Oxyma double couplings. Below we can see the chromatographic differences between the 5-mer cylic and 10-mer cyclic sequences (Figure 2).
For my 10-mer we improved to 72% crude purity of cyclic product compared to ~30% with our 15 mer with little to no dimerization. I did still see linear species which was 10% of my crude material interestingly an increase compared to my 15-mer. This, however, is much more resolved compared to the dimer product and I can work with that in purification.
My 5-mer on the other hand did not perform as I expected. It seems to maintain some dimerization or more likely aggregation. Since I have 2 peaks at ~4.6 minutes it looks like I have two products. However, I observe a single predominant mass in the mass spectrum. The UV chromatogram suggests evidence of a dimer; however, my mass spectrometer does not resolve the isotopic envelope of a mass peak making characterization of the ionization state more difficult. I tried to break aggregate interactions by increasing the temperature of my column from 50C to 60C to and heating my sample to no avail. I also see the wide peak at 5.5 minutes that also seems to be aggregates as there is a large UV signal but there is no distinct mass at any point across this peak in my mass spectrum - just a bunch of noise. What I can see is that my linear product was completely consumed compared to the 10-mer and 15-mer.
Another theory other than aggregation and due to the length of my peptide, cyclization may promote two different conformational states that may be chromatographically resolvable. With bulky side chains and a flexible peptide my termini may come together at different orientations. The macrocycle could be conformationally locked and unable to freely rotate evidenced by the same molecular weight by mass spec but 2 chromatogram peaks. Since I don’t see this peak in my 10-mer I assume there is enough of a difference in sidechains that this reduces the amount of aggregation/conformational locking comparatively to my 5-mer.
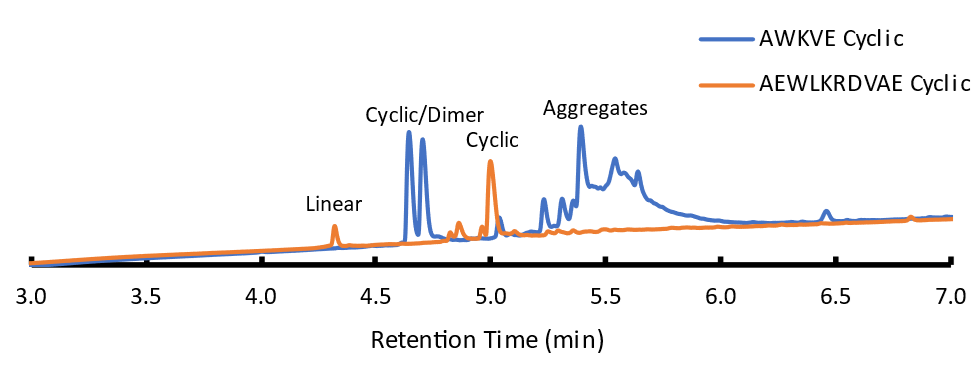
Figure 2: Chromatograms of cyclic products from 5-mer and 10-mer sequences
Overall, there seemed to be a big improvement in cyclization efficiency compared to my 15-mer sequence with significant reduction in dimerization. This most likely could be contributed to the length of my sequence but as always is likely due to the fact that I have different sequences. Are you having success with head-to-tail cyclization? What types of strategies are you using to improve your efficiency. If you want to read more on cyclic peptides check out the links above or if you want to read more about cyclization methods, check out the application note below!
Published: Dec 20, 2023 10:12:06 PM


